Tumor-associated microbiota is an important component of the tumor microenvironment (TME) across 33 types of human cancer. However, little evidence is available regarding the spatial distribution and localization of these microbes in tumor cells.
Addressing this gap in research, a recent Nature journal study evaluated spatial, cellular, and molecular host-microbe interactions in oral squamous cell carcinoma (OSCC) and colorectal cancer (CRC). In this study, scientists mapped host–bacterial cellular, spatial, and molecular interactions within the TME using single-cell RNA sequencing (scRNA-seq) and in situ spatial-profiling technologies.
 Study: Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Image Credit: jovan vitanovski / Shutterstock
Study: Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Image Credit: jovan vitanovski / Shutterstock
Background
Typically, cancer patients’ tumors comprise malignant cells surrounded by a compound network of non-malignant cells. These cells might exhibit pro- or anti-tumorigenic effects based on their abundance and type. Both in vitro and in vivo experiments have indicated the presence of bacteria in the tumor-associated microbiota, which play an important role in cancer development, immunosurveillance, metastasis, and chemoresistance. Molecular analysis and bioimaging data have also shown the existence of intratumoral microbiota across major cancer types.
There is a lack of evidence regarding the specific identity of host cells through which tumor-associated microbes interact with cancer patients’ tumor cells. Additionally, little evidence has been documented related to identifying specific cells that harbor organisms. The effect of precise host–microbial cellular interactions and spatial distribution of the intratumoral microbiota on their functional capabilities within TME is not apparent.
About the Study
16S rRNA gene sequencing of tumor tissues of CRC patients indicated the presence of various bacteria, including Fusobacterium. The abundance of this bacteria differed between CRC patients. Dendrogram analysis and principal component analysis with beta diversity clustering indicated that the majority of cancer patients had relatively stable microbiome compositions. Nevertheless, most patients exhibited varying degrees of heterogeneity in the intratumoral microbiome composition.
The RNAscope–fluorescence in situ hybridization (RNAscope-FISH) imaging confirmed the heterogeneous spatial distribution of bacterial communities in TME. RNAscope-FISH-based data showed the presence of Fusobacterium nucleatum, which was further validated through microbiome analysis and quantitative PCR technique.
10x Visium spatial transcriptomics was also used to detect and analyze the spatial distribution of intratumoral microbiota of CRC and OSCC specimens. This approach identified 28% of captured spots within OSCC tumors and 46% of CRC tumors.
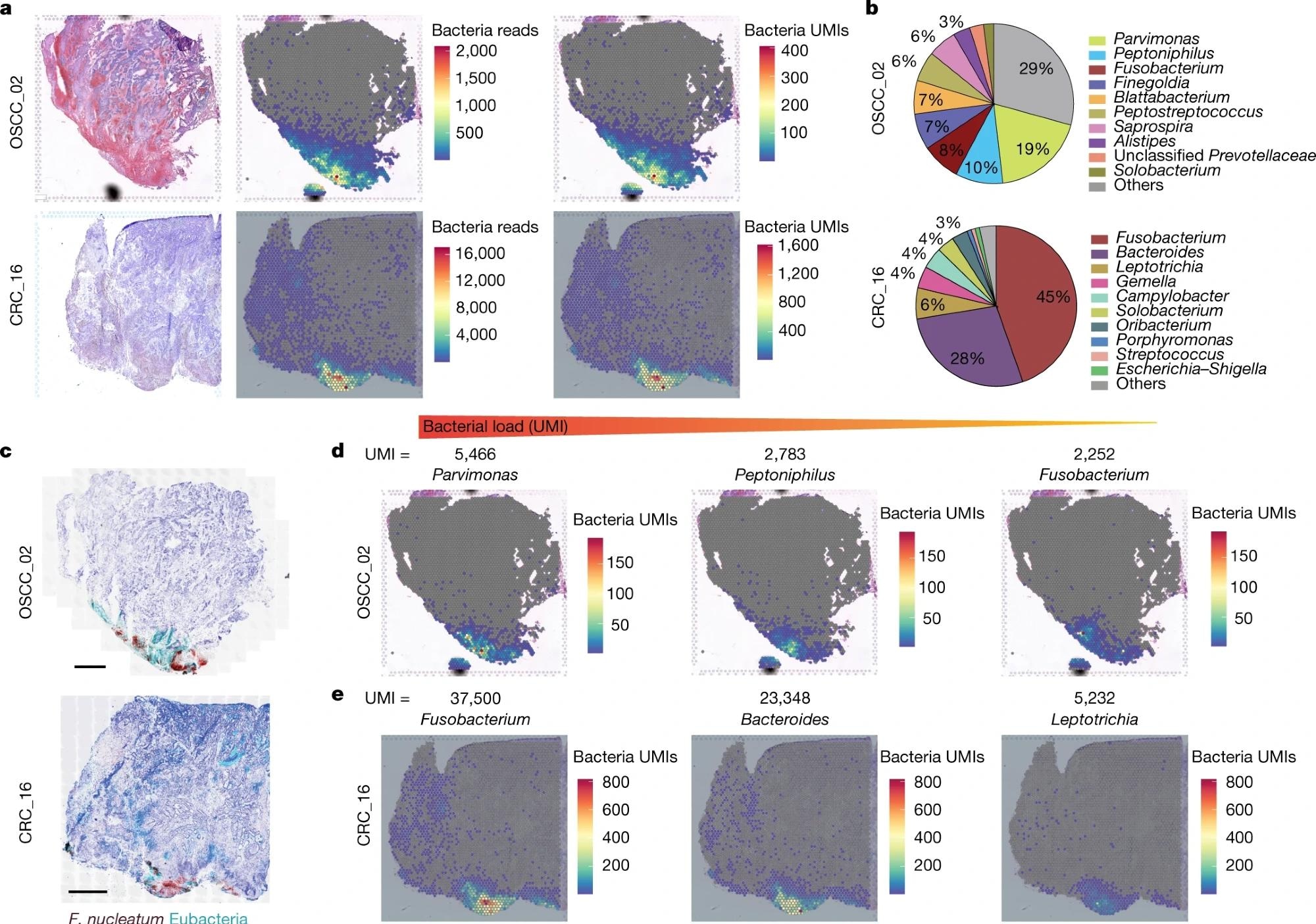 a, Haematoxylin and eosin (H&E) staining (left), spatial distribution of total bacterial reads (centre) and total UMI transcripts (right) throughout the tumour tissue in the 10x Visium capture slides from human OSCC and CRC specimens. b, Pie chart of the top 10 most dominant bacterial genera detected in the 10x Visium RNA-sequencing data from the OSCC and CRC tumours. c, RNAscope-FISH imaging showing the distribution of bacteria across the tumour tissue in a sequential slide following the 10x Visium section. The F. nucleatum probe is red and the eubacterial probe is cyan. Scale bars, 1 mm. d, Spatial distribution of Parvimonas, Peptoniphilus and Fusobacterium UMIs detected in the 10x Visium OSCC specimen data. e, Spatial distribution of Fusobacterium, Bacteroides and Leptotrichia UMIs detected in the 10x Visium CRC specimen data.
a, Haematoxylin and eosin (H&E) staining (left), spatial distribution of total bacterial reads (centre) and total UMI transcripts (right) throughout the tumour tissue in the 10x Visium capture slides from human OSCC and CRC specimens. b, Pie chart of the top 10 most dominant bacterial genera detected in the 10x Visium RNA-sequencing data from the OSCC and CRC tumours. c, RNAscope-FISH imaging showing the distribution of bacteria across the tumour tissue in a sequential slide following the 10x Visium section. The F. nucleatum probe is red and the eubacterial probe is cyan. Scale bars, 1 mm. d, Spatial distribution of Parvimonas, Peptoniphilus and Fusobacterium UMIs detected in the 10x Visium OSCC specimen data. e, Spatial distribution of Fusobacterium, Bacteroides and Leptotrichia UMIs detected in the 10x Visium CRC specimen data.
In the OSCC tumor, Parvimonas, Peptoniphilus, and Fusobacterium were found to be the dominant strains, whereas Fusobacterium and Bacteroides were dominant genera in the CRC tumor.
10x Visium spatial transcriptomics technique enabled direct detection, quantification, and spatially mapping of viable bacteria within cancer patients’ intact tumor tissues. It further indicated the complexity of intratumoral microbiota interactions across tumor tissues.
The GeoMx digital spatial profiling (DSP) platform helped quantify the expression profile of 77 proteins linked with cancer progression and anti-tumor immunity. This technique, combined with RNAscope and the immunohistochemistry (IHC) approach, indicated that bacterial communities populate highly immuno-suppressive microniches and are not much vascularized. Additionally, bacterial strains are prone to inhabit malignant cells with reduced levels of Ki-67.
The INVADEseq (invasion–adhesion-directed expression sequencing) technique was developed to assess bacterial–host cell-to-cell interaction within the TME and the effect on host cell transcriptomics. This technique is associated with the introduction of a primer to target conserved regions of bacterial 16S rRNA. Subsequently, cDNA libraries with bacterial transcripts from the bacteria-associated human cells can be produced. One of the crucial aspects of this method is that the introduction of primer does not affect the gene expression profile of human CRC cells.
The INVADEseq technique was validated using the human CRC cell line HCT116 that was infected with bacterial species, namely, F. nucleatum, Porphyromonas gingivalis, and Prevotella intermedia. It enabled the mapping of bacterial populations in single human cells. Importantly, INVADEseq confirmed the role of F. nucleatum and P. gingivalis in affecting cancer cell heterogeneity. These bacterial strains alter distinct transcriptional programs that aid in specific cell clustering. Alteration of transcriptional pathways is also associated with the manifestation of inflammation, cell dormancy, metastasis, and DNA repair.
Conclusions
The present study revealed that cancer cells infected with bacteria affect their surrounding as a single cell, which subsequently employs myeloid cells to the bacterial territory. Notably, the microbiome within the tumor was found not to be a random phenomenon. Instead, it was stated that the presence of bacteria within a tumor is a highly organized process in microniches with immune and epithelial cell functions that influence cancer progression.
Even though the current study focussed on two cancer types, the tools and techniques could be used to study all major types of cancer and those containing intratumoral microbiota.