In a recent research letter published in the journal Nature Aging, researchers used proteomic sequencing of patients’ and controls’ cerebrospinal fluids to investigate the heterogeneity of Alzheimer’s disease. Their findings revealed five molecular subtypes that depicted distinct genetic risk factors and disease pathologies, including progression rates and survival times. These results suggest different intervention requirements for each subtype and highlight the need for personalized medicine to diagnose and treat the condition.
 Letter: Cerebrospinal fluid proteomics in patients with Alzheimer’s disease reveals five molecular subtypes with distinct genetic risk profiles. Image Credit: Lightspring / Shutterstock
Letter: Cerebrospinal fluid proteomics in patients with Alzheimer’s disease reveals five molecular subtypes with distinct genetic risk profiles. Image Credit: Lightspring / Shutterstock
Alzheimer’s disease and the advantages of proteomics
Alzheimer’s disease (AD) is a progressive brain disorder predominantly affecting aged individuals, characterized by the degeneration of neurons responsible for memory and cognition. It is estimated to affect 5% of individuals between the ages of 65-74, 13.1% between 75-84, and 33.3% above 84 years, currently affecting 44 million people, with this number rising annually. AD is recognized as the leading cause of dementia worldwide, with no cure presently known and treatment restricted to symptom management. While a definitive underpinning for the disease is yet to be identified, genetics and environmental exposure are assumed responsible for the condition.
Recent research has identified that AD is not a single disease but an umbrella term for a spectrum of conditions that vary significantly at the molecular level. Unfortunately, these research advancements invalidate a large body of previous literature attempting to elucidate the clinical pathophysiology of AD, given that different patients may respond significantly differently to the same clinical exposure.
‘Proteomics’ is the study of the interactions, function, composition, and structures of proteins and their cellular activities. It incorporates cutting-edge ‘next-generation’ sequencing techniques such as mass spectrometry (MS) to identify and characterize thousands of protein subunits in biofluids. Cerebrospinal fluid (CSF) is the most accessible of these biofluids relating to neurological conditions due to its constant contact with the brain and central nervous system (CNS) and its role as a proxy for the brain’s pathophysiological process.
About the study
In the present study, researchers used a case-control cohort approach, using CSF from AD patients and age-matched healthy controls, to reveal the differentially up- and down-regulated proteins in these cohorts via proteomic analyses. The study sample group was derived from the Amsterdam Dementia Cohort (ADC), an ongoing study of all patients who have sought treatment at the Alzheimer’s Centre in Amsterdam since 2000.
Study inclusion criteria comprised diagnosed AD, confirmed based on the presence of an abnormal amyloid marker (cases) and age, sex, and demographic-matched controls. CSF from both cohorts was collected and subjected to high-performance liquid chromatography (HPLC) mass spectrometry (MS) – LC-MS/MS. Enzyme-linked immunosorbent assays (ELISAs) were then used to measure Amyloid-β42, t-tau, p-tau 181, and the amyloid-β42/amyloid-β40 ratio, the main determinants of AD severity and progression stage.
Blood samples from cases and controls were further subjected to apolipoprotein E (APOE) genotyping to screen for single-nucleotide polymorphisms that are known to enhance or suppress AD. T1-weighted magnetic resonance imaging (MRI) was used to visualize brain atrophy patterns and evaluate the differences in AD patients’ and controls’ neuroimages. Finally, standardized neuropsychological test batteries were administered to study subjects during initial enrollment, with annual follow-up to estimate the rate and degree of AD progression.
Study findings
The present study included 609 cases and 187 controls. Of the included AD cases, 107 displayed normal cognition, 103 displayed mild cognitive impairment (MCI), and 209 displayed dementia. LC-MS/MS analyses identified 3,863 unique CSF proteins, of which 1,309 proteins (28,408 peptides) were common to all included participants and were used for further analyses. Of these, cluster analyses revealed 1,058 AD-related proteins. Combining clustering results with patients’ clinical characteristics revealed five distinct AD subtypes.
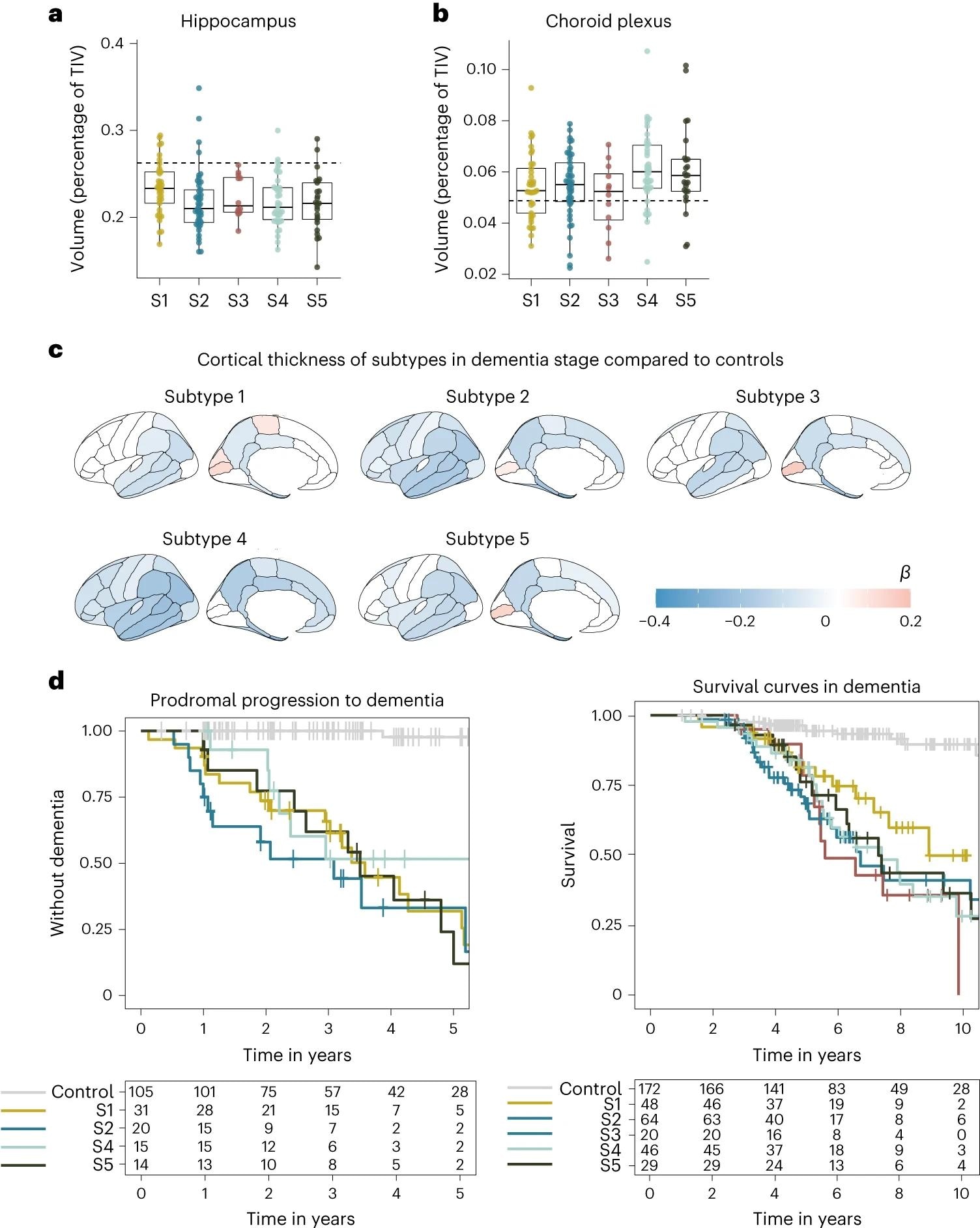 a, Median hippocampal volume as the percentage of total intracranial volume (TIV) compared to subtypes in the dementia stage. b, Choroid plexus volume as the percentage of TIV compared to subtypes in the dementia stage. c, Cortical atrophy associated with AD subtypes in the dementia stage compared to controls (n = 160). β indicates mean cortical thickness in mm, averaged over the right and left hemispheres and adjusted for age and sex. d, Clinical progression from MCI to dementia according to subtype (left; excluding subtype 3 due to n = 2) and time from dementia to death according to subtypes (right). All atrophy measures are based on individuals with dementia only. a,b, The boxplots depict the median in the center; the boundaries indicate the first and third quartiles, while the whiskers extend up and down to 1.5 times the interquartile range (limited to actual observed data points), and the points indicate individual person values (subtype 1, n = 37; subtype 2, n = 45; subtype 3, n = 12; subtype 4, n = 40; subtype 5, n = 25).
a, Median hippocampal volume as the percentage of total intracranial volume (TIV) compared to subtypes in the dementia stage. b, Choroid plexus volume as the percentage of TIV compared to subtypes in the dementia stage. c, Cortical atrophy associated with AD subtypes in the dementia stage compared to controls (n = 160). β indicates mean cortical thickness in mm, averaged over the right and left hemispheres and adjusted for age and sex. d, Clinical progression from MCI to dementia according to subtype (left; excluding subtype 3 due to n = 2) and time from dementia to death according to subtypes (right). All atrophy measures are based on individuals with dementia only. a,b, The boxplots depict the median in the center; the boundaries indicate the first and third quartiles, while the whiskers extend up and down to 1.5 times the interquartile range (limited to actual observed data points), and the points indicate individual person values (subtype 1, n = 37; subtype 2, n = 45; subtype 3, n = 12; subtype 4, n = 40; subtype 5, n = 25).
Subtype 1 is characterized by neuronal hyperplasticity, subtype 2 by innate immune activation, subtype 3 by RNA dysregulation, subtype 4 by choroid plexus dysfunction, and subtype 5 by blood-brain barrier dysfunction. APOE genotyping corroborated identified clusters and suggested a unique genetic underpinning for each subtype.
“Notably, we found that each subtype was associated with distinct AD genetic risk factors, further supporting that each CSF AD subtype reflects specific underlying molecular mechanisms. The subtypes also differed in cortical atrophy patterns and survival times, underscoring their clinical relevance.”
Subtypes were found to differ significantly by their clinical pathology, as highlighted by neurophysiological testing – subtype 3 was substantially more aggressive in its progression rate compared to the other subtypes. Given the degree of genetic and pathophysiological uniqueness of these subtypes, the need for personalized medicine becomes apparent.
“…side effects arising from certain treatments may also depend on subtype. For example, while antibodies may more easily cross the blood–brain barrier in subtype 5, these individuals may be at increased risk for cerebral bleeding that can occur with antibody treatment.”
Conclusions
The present study used proteomics to investigate the patient-specific differences in genetic and pathophysiological profiles under the AD umbrella. Study findings reveal more than 1,000 proteins differentially expressed in AD patients, and importantly, that AD comprises at least five distinct subtypes differing in their genetic and clinical underpinnings.
“Given the distinct patterns of molecular processes and AD genetic risk profiles, it is likely that AD subtypes will require specific treatments. For example, subtype 1 individuals may benefit from TREM2-activating treatments, subtype 2 from innate immune inhibitors, subtype 3 from antisense oligonucleotides that restore RNA processing, subtype 4 from inhibition of monocyte infiltration and subtype 5 from cerebrovascular treatments.”
Journal reference:
- Tijms, B. M., Vromen, E. M., Mjaavatten, O., Holstege, H., Reus, L. M., Wesenhagen, K. E., Lorenzini, L., Vermunt, L., Venkatraghavan, V., Tesi, N., Tomassen, J., Den Braber, A., Goossens, J., Vanmechelen, E., Barkhof, F., Pijnenburg, Y. A., M., W., Teunissen, C. E., Berven, F. S., . . . Visser, P. J. (2024). Cerebrospinal fluid proteomics in patients with Alzheimer’s disease reveals five molecular subtypes with distinct genetic risk profiles. Nature Aging, 1-15, DOI – https://doi.org/10.1038/s43587-023-00550-7, https://www.nature.com/articles/s43587-023-00550-7