In a recent article published in Nature Communications, researchers performed a genomic investigation of oral resistome development, i.e., of genes that confer antimicrobial resistance (AMR).

Background
Since AMR is a growing health and economic issue, characterizing the AMR of the human oral microbiome is of utmost importance. After the gut, the oral cavity houses the most microbes within the human body.
Accordingly, researchers have identified antimicrobial resistance genes (ARGs) in the oral microbiome of neonates to adults. Moreover, the oral microbiome is a well-recognized site where horizontal gene transfer (HGT) occurs, which, in turn, facilitates the development of antimicrobial-resistant infections.
In childhood, diet, the introduction of solid foods, and the emergence of teeth change the composition of the oral microbiome. Host genetics also comes into play and influences ARG-carrying oral bacteria during childhood. However, it has remained unexplored how commensal and pathogenic bacteria in the oral microbiome acquire and develop AMR during childhood.
One Health recognizes the connectedness of human and animal health as they share the same environment. Since the oral microbiome is a significant antimicrobial resistance (AMR) reservoir, its surveillance, as part of the One Health Approach, is necessary to combat AMR spread due to antibiotic overuse.
About the study
Oral microbiome diversity increases as children grow older. So, in the present study, researchers sequentially examined 530 oral metagenomes from 221 Australian twins and demonstrated the widespread presence of ARGs in their oral microbiome. They examined the development of the oral resistome, including taxonomic and functional association, in addition to the mobilization potential of ARGs.
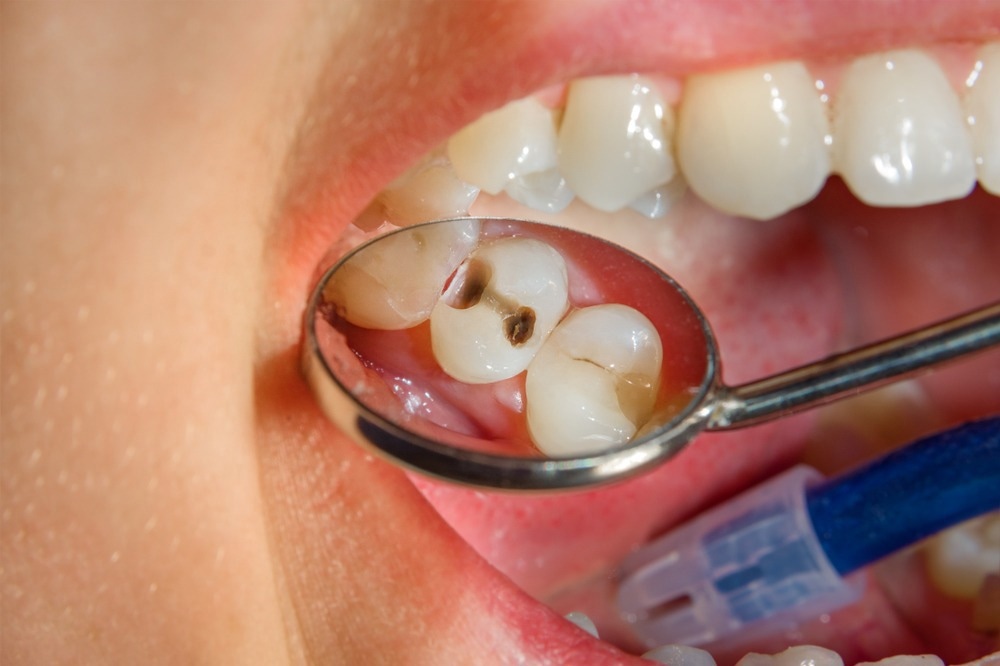
In addition, they showed how it significantly changed in composition with other microbiome constituents over the first decade of life and in response to changes in oral health, e.g., dental caries and placement of restorations.
The twin study design allowed easy comparison of varying AMR phenotypes of monozygotic (MZ) and dizygotic (DZ) twins. More importantly, it helped the researchers take a sneak peek into how genetics and environment influence the development of the oral resistome in children aged between 2.4 months and 10.8 years.
Results
Of the 221 twins, 124 and 97 were females and males children, respectively. The study results showed that the oral resistome of these children played a significant role in AMR imbalance and transmission. It was also inherently dynamic.
Although scientists have observed that oral resistome accounts for less than 1% of the known microbiome, in this study, they observed that AMR-related mobile genetic elements (MGEs) were extensively prevalent, for instance, the Tn916 transposase family. Another remarkable finding was that the mobilization potential of ARGs increased with the age of children. Moreover, genetics and environment, e.g., early feeding practices, influenced the oral resistome composition, just like they influenced the oral microbiome. Furthermore, oral health altered the resistome composition.
The longitudinal profiling of the oral metagenome also showed that the oral resistome was stable and returned to equilibrium after short-term perturbations. Its resilience remained unaffected by significant disturbances in the oral cavity during the first two and a half years of life due to tooth eruption and dietary changes. However, oral resistome exhibited temporary changes in diversity before permanently stabilizing at the age of five years (T3). On the contrary, gut resistome substantially increases its ARG richness only within the first year of life.
Functional investigations revealed that after T3, ARGs interacted with functional pathways, e.g., the mycothiol biosynthesis pathway, to facilitate the development of AMR. Mycothiol detoxifies antibiotics, enabling good bacteria to endure antibiotic exposure and eventually develop resistance. Individuals with higher ARG diversity also showed a higher potential for biofilm building, e.g., sugar degradation pathways. In the future, transcriptomics-based studies could explain the association between AMR and bacterial metabolism.
Taxonomic investigations of the oral resistome revealed the spread of AMR via HGT co-location of ARGs and insertion sequences (IS) in 27 species across varying time points. They observed that oral commensals, such as Streptococcus mitis and S. anginosus carried IS-associated ARGs. These species were exceptionally resistant to treatment and passed resistance to new environments they entered. These species exhibited an association with the Tn916 family of transposases and facilitated the co-carriage of two resistance genes, tetracycline resistance gene tet(m) and macrolide resistance gene erm(B).
The authors could determine the pattern of heritability reliably for T2 and T3 only, not T1. The heritability effect declined between T2 and T3. In contrast, the influence of standard and unique environmental effects increased on the oral resistome, likely because compared to infants, school-goers experience diverse environments, e.g., diet and antibiotics. Furthermore, the researchers noted that though indirect exposures, e.g., antibiotic use in food production, influenced oral resistome composition, protein consumption did not significantly affect resistome diversity.
Conclusions
To conclude, the study highlighted the importance of understanding how the oral resistome and its interactions with commensals and other bacterial species, restorative materials, etc., is critical to improving oral health. Since the connections between the oral cavity and respiratory, vascular, and digestive systems are intimate, mobilization of the oral resistome might have a long-term impact on systemic health beyond childhood.