A large number of health-related benefits of probiotics have led to the development of a new discipline called probiogenomics, which focuses on the molecular mechanisms of probiotic activities. According to the US Food and Drug Administration (FDA), the next-generation probiotics satisfy the scope of “live biotherapeutic products” (LBPs), which could be used to develop effective therapeutics for various diseases.
 Study: The Human Milk Microbiota Produces Potential Therapeutic Biomolecules and Shapes the Intestinal Microbiota of Infants. Image Credit: OlgaReukova / Shutterstock
Study: The Human Milk Microbiota Produces Potential Therapeutic Biomolecules and Shapes the Intestinal Microbiota of Infants. Image Credit: OlgaReukova / Shutterstock
Background
The combination of next-generation probiotics with omics technologies has immensely progressed the application of probiotics. A wide range of biomolecules could trigger the multifactorial molecular mechanisms related to the health benefits of probiotics. These biomolecules are mostly metabolic products of microbes or protein molecules present on the cell surface (e.g., Lactobacillus surface (S)-layer proteins).
In Lactobacillus, the S protein covers the entire bacterial cell. It forms a layer that protects from osmotic and mechanical stress, bacteriolytic enzymes, changes in pH values, and antimicrobial peptides. Only a few Lactobacillus strains produce S-proteins that play an essential role in the expression of probiotic properties. This S-layer is in direct contact with the microenvironment and assists in the adhesion to mucin and epithelial cells, mediating surface recognition, autoaggregation, and coaggregation.
Bioactive peptides, exopolysaccharides (EPSs), and bacteriocins are potential biomolecules with health-promoting effects. Bacteriocins contain antimicrobial properties that help probiotic cells to withstand a competitive polymicrobial environment. EPS plays a protective role when probiotic cells are exposed to the intestinal environment. Additionally, this biomolecule exerts a bifidogenic effect on intestinal microbiota and drives immunomodulatory, antimicrobial, antitumor, and cholesterol-lowering activities.
Biomolecules are also known as signaling molecules that can detect microbial interactions as well as regulate signaling pathways. In addition, biologically active molecules are applied as biotherapeutics, particularly as an alternative to antibiotics.
A key source of novel probiotic strains is the human milk microbiome, which constitutes bacterial species transferred during pregnancy and birth. The human milk microbiome positively helps establish the intestinal microbiota of newborns.
About the Study
A recent International Journal of Molecular Sciences study evaluated the contribution of the human milk microbiota in shaping an infant’s intestinal microbiota. Additionally, unique bacterial isolates were identified from the human milk samples obtained from Croatian mothers. The authors also characterized specific cellular molecules, especially, S-layer proteins, bacteriocins, and EPSs, which could be used as probiotics in the future. These biomolecules also possess biotherapeutic properties.
This study recruited healthy mothers who breastfed their newborns for at least one month after introducing solid food. Fifteen human milk and infant feces samples were collected separately at three distinct points, i.e., within one week of the infant’s birth, one month after birth, and one month after the introduction of solid food. Around 100 bacterial strains were isolated from human milk samples.
Lactobacillus helveticus, an S-layer expressing strain, was used as a positive control, whereas Lactiplantibacillus plantarum, a non-S-layer producing strain, was used as a negative control. Antimicrobial properties were assessed against Listeria monocytogenes, Escherichia coli, Staphylococcus aureus, and Salmonella enterica.
Study Findings
MiSeq sequencing data confirmed that human milk and infant feces samples had a unique microbiome composition. The composition of the human milk microbiome varied among mothers. For instance, the abundance of Firmicutes varied between 43.36% and 83.37%, Bacteroidetes from 0.17% to 13.94%, Actinobacteria from 0.46% to 21.89%, Proteobacteria from 2.86% to 53.46% and Verrucomicrobia from 0% to 7.38%. In infants, the fecal microbiome contained a varied level of Firmicutes, Proteobacteria, Actinobacteria, Veruccomicrobia, and Bacteroidetes.
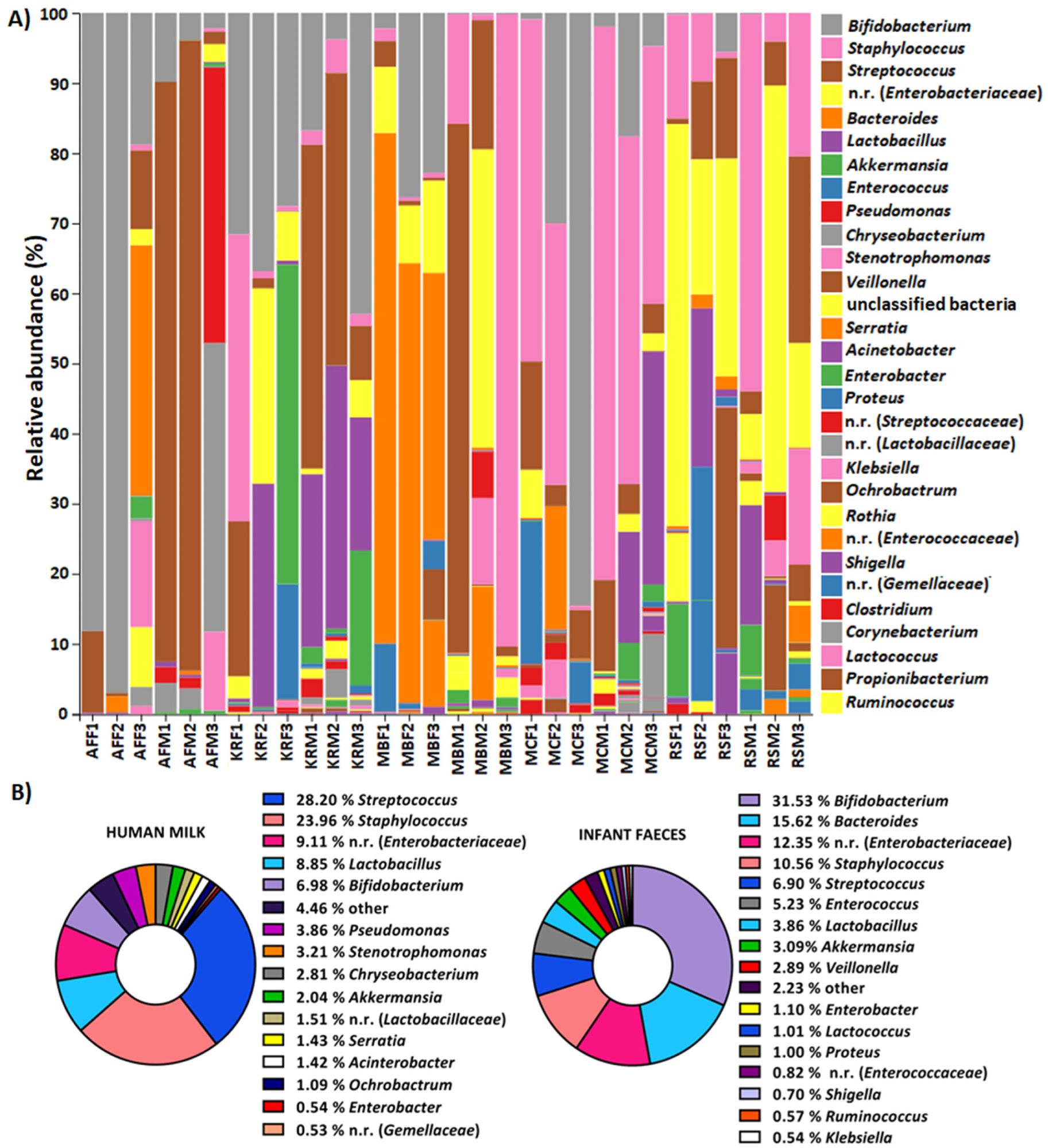 Relative abundance of OTUs at genus level of: (A) human milk samples from the same mother (M), i.e., feces (F) from the same infant, (B) all human milk and infant feces samples. OTUs with an average representation of less than 0.5% are classified as other. n.r.—not recognized genus from the family listed in parentheses.
Relative abundance of OTUs at genus level of: (A) human milk samples from the same mother (M), i.e., feces (F) from the same infant, (B) all human milk and infant feces samples. OTUs with an average representation of less than 0.5% are classified as other. n.r.—not recognized genus from the family listed in parentheses.
While human breastmilk and infants’ feces microbiomes were similar, human breastmilk exhibited a relatively higher abundance of the same phyla than feces. In addition, the α-diversity of the milk microbiome was found to be significantly higher compared to feces. This finding indicates that the milk microbiota is more diverse than the fecal microbiota.
A change in the human milk microbiome composition was observed at different lactation periods, which affected the infant’s fecal microbiome composition. For instance, over time, the amount of Actinobacteria and Verrucomicrobia was enhanced in human milk and the infant’s feces. Notably, after the introduction of solid food, bacteria belonging to the phylum Bacteroidetes increased substantially.
A total of 28 genetically distinct strains of bacteria, which are producers of therapeutic biomolecules, were selected using hierarchical cluster analysis of RAPD-PCR electrophoretic profiles. Four L. brevis strains were isolated from the human milk microbiome that synthesized S-layer proteins and demonstrated immunomodulatory activity in the gut. Additionally, six L. plantarum strains produced plantaricin, which conferred an antibacterial effect. Only L. fermentum was identified to synthesize EPS.
Conclusions
The current study revealed that each human milk and infant’s feces sample is unique concerning the microbiome. However, both the microbiomes studied were found to be producers of biomolecules possessing therapeutic effects. In the future, the authors will further analyze the bacterial isolates and their potential therapeutic biomolecules. These biomolecules could be potential next-generation probiotics.